2.1 Introduction
Describing the topology of networks of seed circulation is interesting since it gives insight on how exchanges are organized within a PPB programme or a Community Seed Bank(Vernooy, Shrestha, and Sthapit 2015)(Pautasso et al. 2013). Analysis can be done at several geographical or organizing scales, for example local, regional or national.
Two types of network are handled within PPBstats :
- unipart network where nodes are:
- seed lots (i.e. a combinaison of a germplasm in a given location a given year) and edges are relationships such as diffusion, mixture, reproduction, crosses or selection for example.
- locations and edges are diffusion events between locations
- bipart network where nodes are either location or germplasm
2.1.1 Workflow and function relations in PPBstats regarding network analysis
The workflow is very simple as only descriptive analysis can be done based on network format (Figure 2.1).
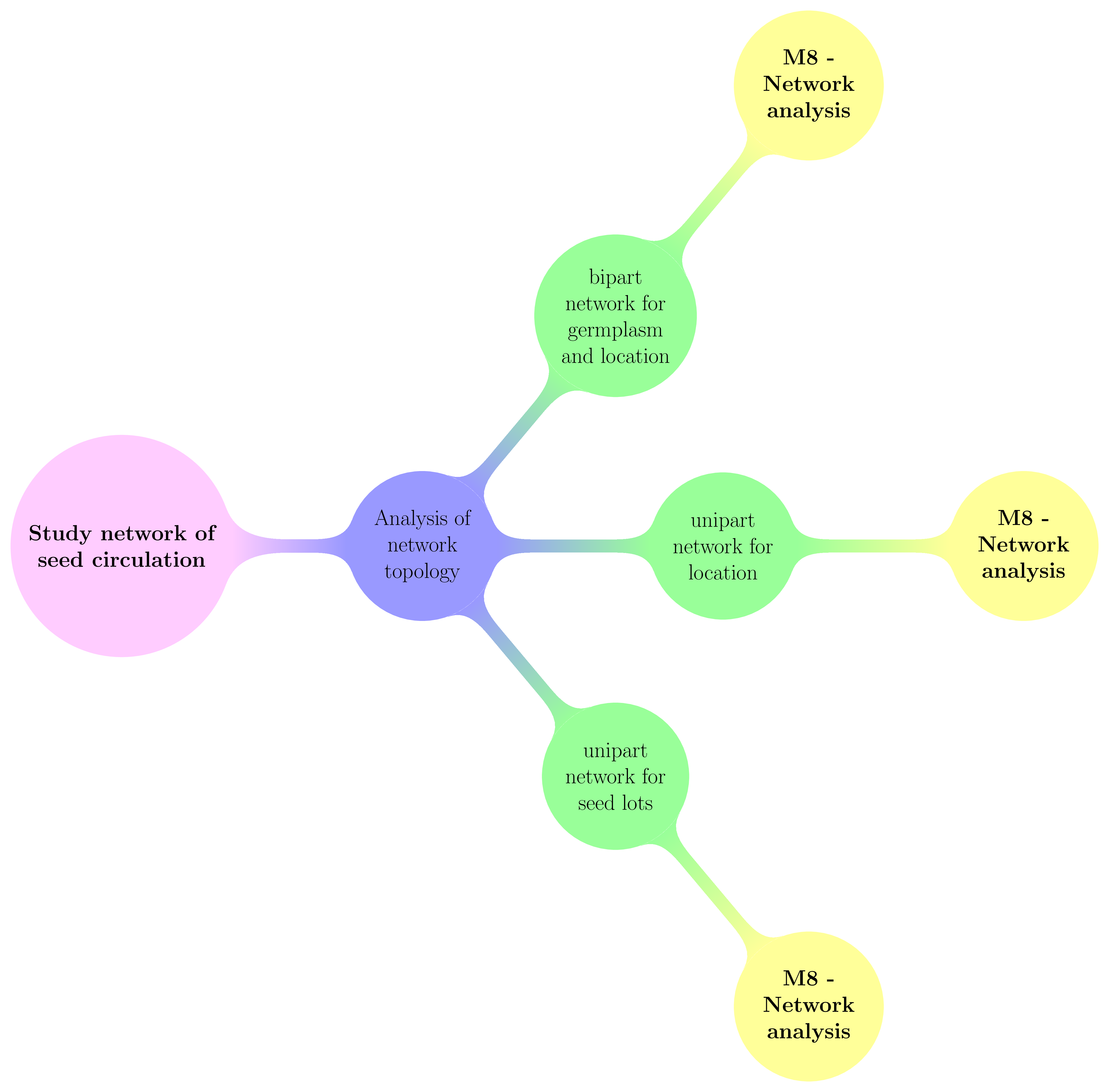
Figure 2.1: Decision tree with objectives and analysis carry out in PPBstats regarding network analysis. M refers to methods.
Figure 2.2 displays the functions and their relationships. Table 2.1 describes each of the functions.
You can have more information for each function by typing ?function_name in your R session.
Note that plot() is S3 method.
Therefore, you should type ?plot.PPBstats to have general features and then see in details for specific functions.
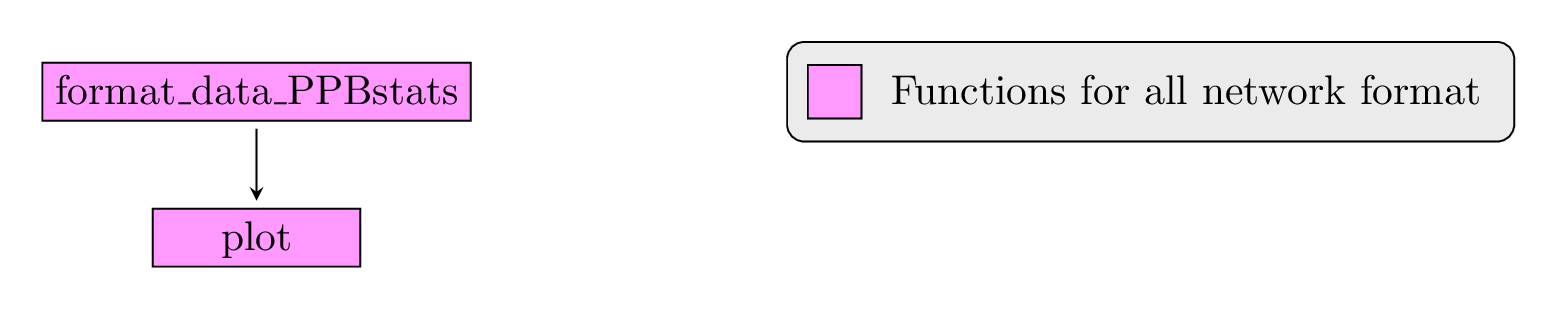
Figure 2.2: Main functions used in the workflow.
| function name | description |
|---|---|
format_data_PPBstats |
Check and format the data to be used in PPBstats functions |
plot |
Build ggplot objects to visualize output |
References
Pautasso, M., G. Aistara, A. Barnaud, S. Caillon, P. Clouvel, O. Coomes, M. Delêtre, et al. 2013. “Seed exchange networks for agrobiodiversity conservation. A review.” Agronomy for Sustainable Development 33.
Vernooy, R., P. Shrestha, and B. Sthapit. 2015. Community Seed Banks: Origins, Evolution and Prospects. Issues in Agricultural Biodiversity. Earthscan for Routledge.