3.2 Design the experiment
The experimental design is described by the number of plots per location, the number of locations, the number of replications of the different germplasms within and between locations.
Function design_experiment sets the experimental design based on :
- the number of entries
- the number of controls per block
- the number of blocks
- the number of columns in the design. The number of rows is computed automaticaly.
The function returns a list with
- a data frame
- an image of the experimental design
A description of the algorithm is describe in the help of the function: ?design_experiment.
The four experimental designs are
- D1 - fully-repicated
- D2 - incomplete block design
- D3 - row-colum
- D4 - satellite-farm and regional-farm
3.2.1 Fully-replicated (D1)
In fully replicated design, all entries are replicated with randomly choosen order into different blocks.
The decision tree where fully-replicated designs are used is displayed in Figure 3.3.
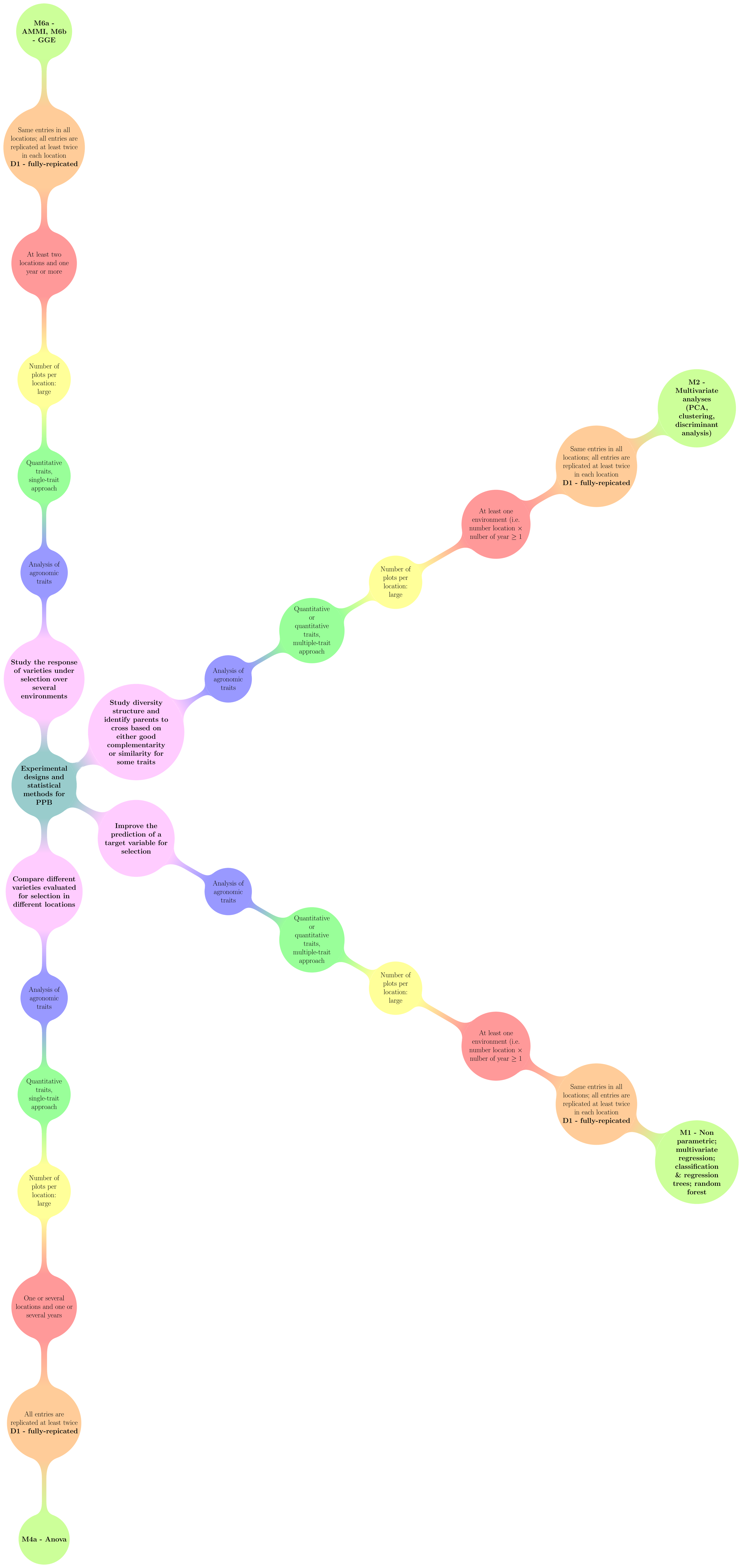
Figure 3.3: The decision tree where fully-replicated designs are used.
The R code to implement is the following :
p_fr = design_experiment(
location = "Location-1",
year = "2016",
expe.type = "fully-replicated",
germplasm = paste("germ", c(1:20), sep = ":"),
nb.blocks = 3,
nb.cols = 4)## Warning in get_data.frame(location, year, nb.germplasm, nb.blocks,
## nb.controls.per.block, : Germplasm are missing to get complete design,
## therefore XXXX are placed in order to fill the design.By default, the data frame is under a standard format:
head(p_fr$"fully-replicated"$data.frame)## location year germplasm block X Y
## 1 Location-1 2016 germ:1 1 A 1
## 2 Location-1 2016 germ:5 1 A 2
## 3 Location-1 2016 germ:13 1 A 3
## 4 Location-1 2016 germ:17 1 A 4
## 5 Location-1 2016 germ:9 1 A 5
## 6 Location-1 2016 germ:4 1 A 6You can set the format to a SHiNeMaS^[Seeds History and Network Management System, see here for more details and a presentation in french here reproduction template file:
p_fr = design_experiment(
location = "Location-2",
year = "2016",
expe.type = "fully-replicated",
germplasm = paste("germ", c(1:20), sep = ":"),
nb.blocks = 3,
nb.cols = 4,
return.format = "shinemas")## Warning in get_data.frame(location, year, nb.germplasm, nb.blocks,
## nb.controls.per.block, : Germplasm are missing to get complete design,
## therefore XXXX are placed in order to fill the design.head(p_fr$"fully-replicated"$data.frame)## project sown_year harvested_year id_seed_lot_sown
## 1 2016 germ:20_Location-2_2016
## 2 2016 germ:5_Location-2_2016
## 3 2016 germ:4_Location-2_2016
## 4 2016 germ:11_Location-2_2016
## 5 2016 germ:9_Location-2_2016
## 6 2016 germ:18_Location-2_2016
## intra_selection_name etiquette split quantity_sown quantity_harvested
## 1
## 2
## 3
## 4
## 5
## 6
## block X Y
## 1 1 A 1
## 2 1 A 2
## 3 1 A 3
## 4 1 A 4
## 5 1 A 5
## 6 1 A 6p_fr$"fully-replicated"$design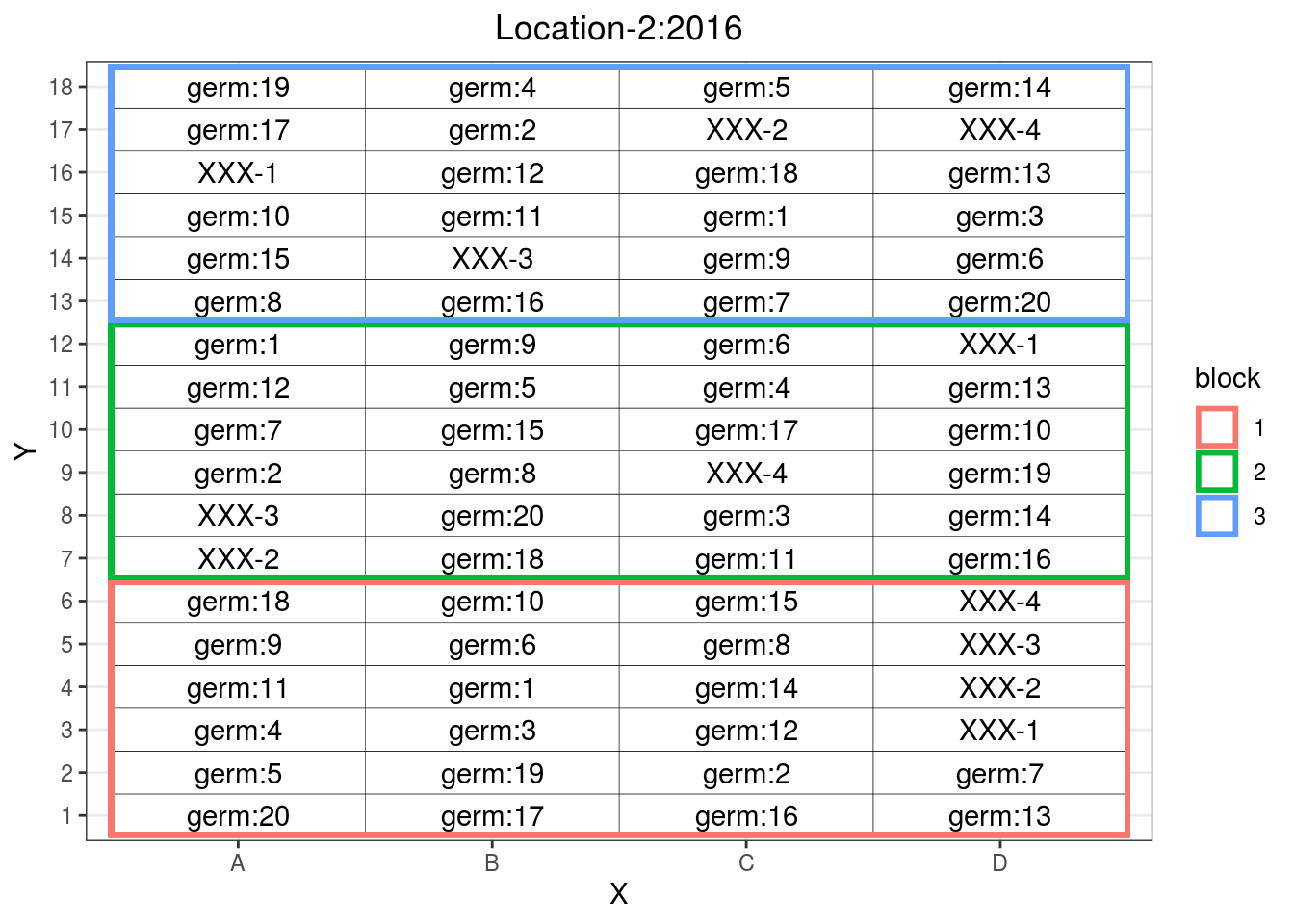
Figure 3.4: Fully replicated design where all germplasms are replicated three time in bocks
3.2.2 Incomplete Block Design (D2)
The decision tree where incomplete block designs are used is displayed in Figure 3.5.

Figure 3.5: The decision tree where incomplete block designs are used.
Entries are not replicated on a location. Some entries are common to some locations. Each block is an independent unit and can be allocated to any location. Each farmer has to choose one or several pre-designed blocks. Therefore, the experiment can be handled by several locations that can not receive a high number of plots.
p_ibd = design_experiment(
year = "2016",
expe.type = "IBD",
germplasm = paste("germ", c(1:10), sep = ":"),
nb.blocks = 8, # i.e. number of location if each location has one block
nb.cols = 4)head(p_ibd$`IBD`$data.frame)## germplasm block X Y year
## 1 germ:1 1 A 1 2016
## 2 germ:1 2 A 2 2016
## 3 germ:5 3 A 3 2016
## 4 germ:3 4 A 4 2016
## 5 germ:2 5 A 5 2016
## 6 germ:3 6 A 6 2016p_ibd$`IBD`$design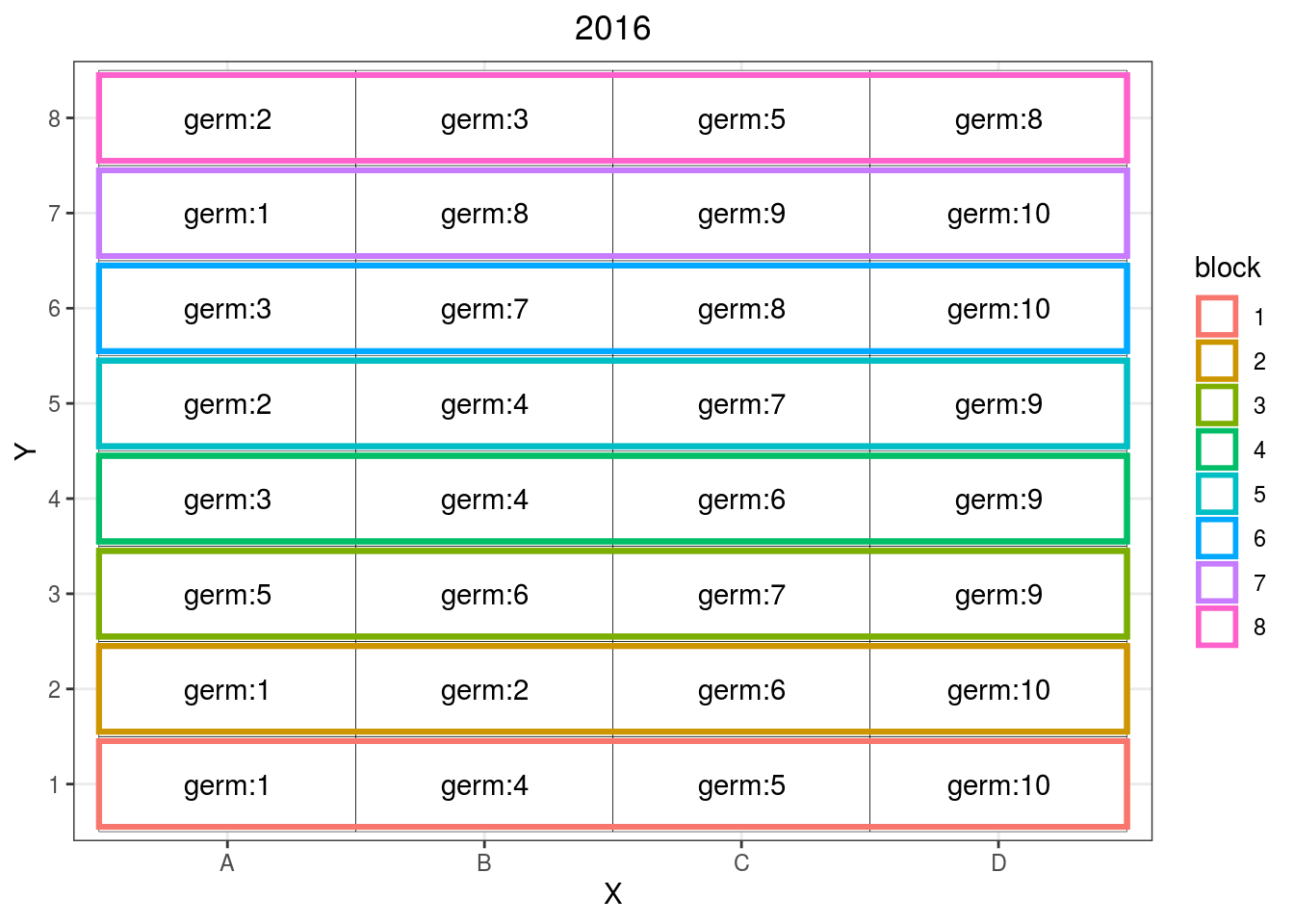
Figure 3.6: Example of incomplete block design where different germplasms are replicated over some blocks.
3.2.3 Row-column (D3)
In Row-column design, a control is replicated in row and columns to catch as much as possible of the variation.
The decision tree where row-column design are used is displayed in Figure 3.7.
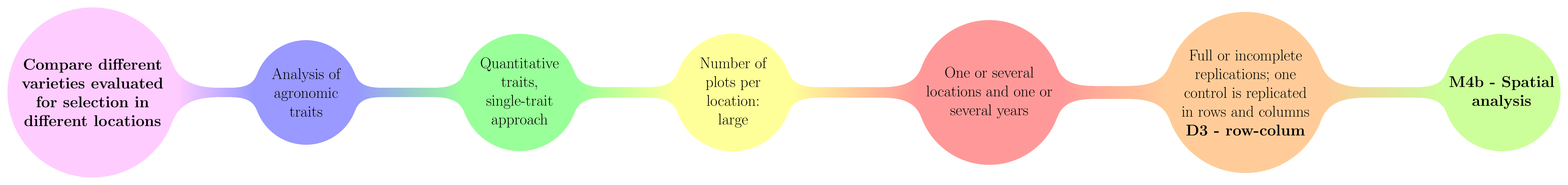
Figure 3.7: The decision tree where row-column designs are used.
The R code to implement is the following :
p_case2 = design_experiment(
location = "Location-3",
year = "2016",
expe.type = "row-column",
germplasm = paste("germ", c(1:20), sep = ":"),
controls = "germ:ctrl",
nb.controls.per.block = 7,
nb.blocks = 1,
nb.cols = 7)head(p_case2$"row-column"$data.frame)## location year germplasm block X Y
## 1 Location-3 2016 germ:5 1 A 1
## 2 Location-3 2016 germ:7 1 A 2
## 3 Location-3 2016 germ:ctrl 1 A 3
## 4 Location-3 2016 germ:20 1 A 4
## 5 Location-3 2016 germ:17 1 B 1
## 6 Location-3 2016 germ:ctrl 1 B 2p_case2$"row-column"$design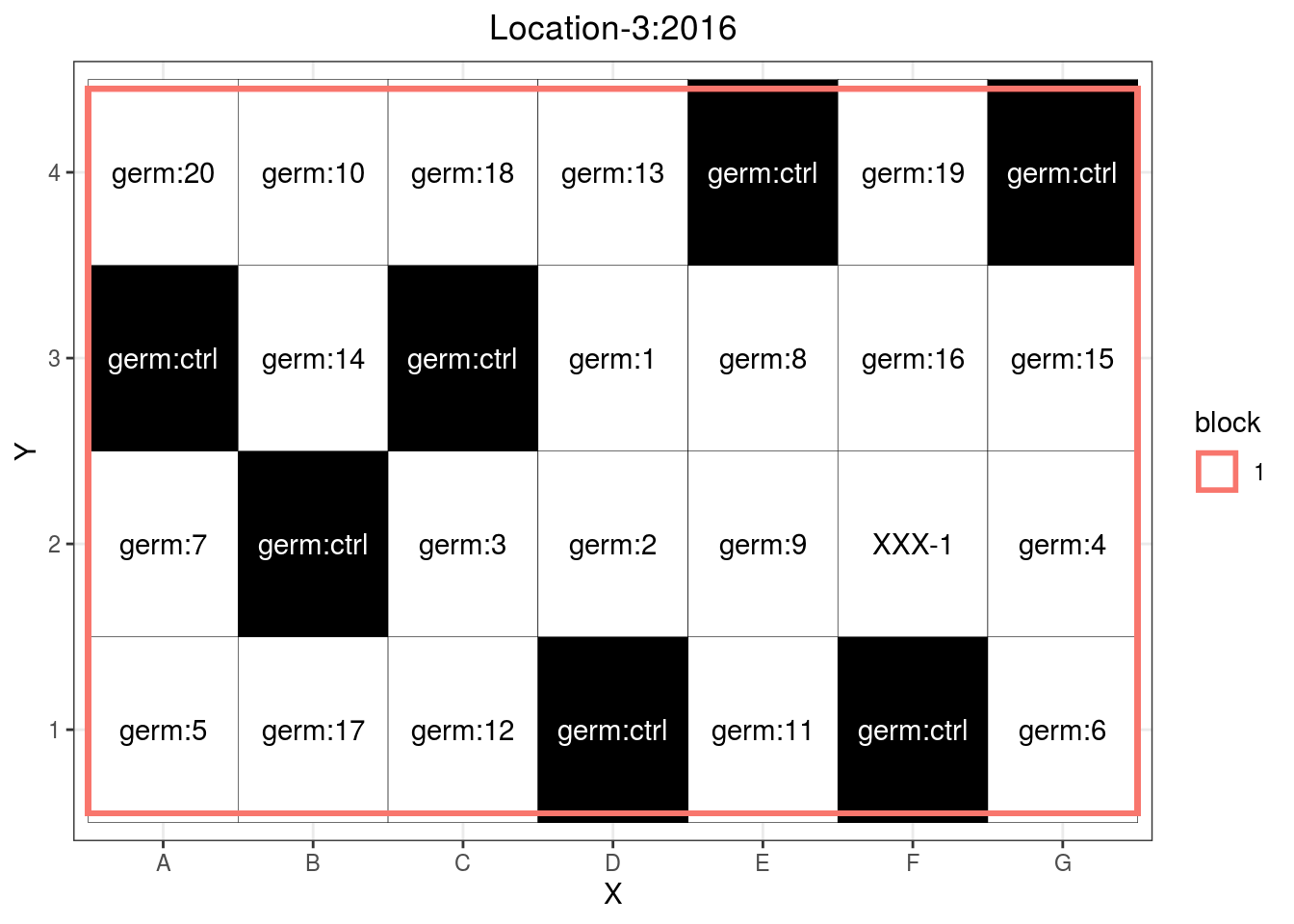
Figure 3.8: Row column design where a control (germ:ctrl) is replicated in rows and columns.
Note that if controls are missing in rows or columns, the function will return an error message. The controls must catch as much as possible of the trial variation.
3.2.4 Regional and satellite farms (D4)
Regional farms receive several entries (i.e. a germplasm in an environment) in two or more blocks with some entries (i.e. controls) replicated in each block. Satellite farms have a single block and only one entry (i.e. the control) is replicated twice. Farmers choose all entries that are not replicated. The number of entries may vary between farms. Note that at least 25 environments (location \(\times\) year) are need in order to get robust results.
The decision tree where regional and satellite farms designs are used is displayed in Figure 3.9.
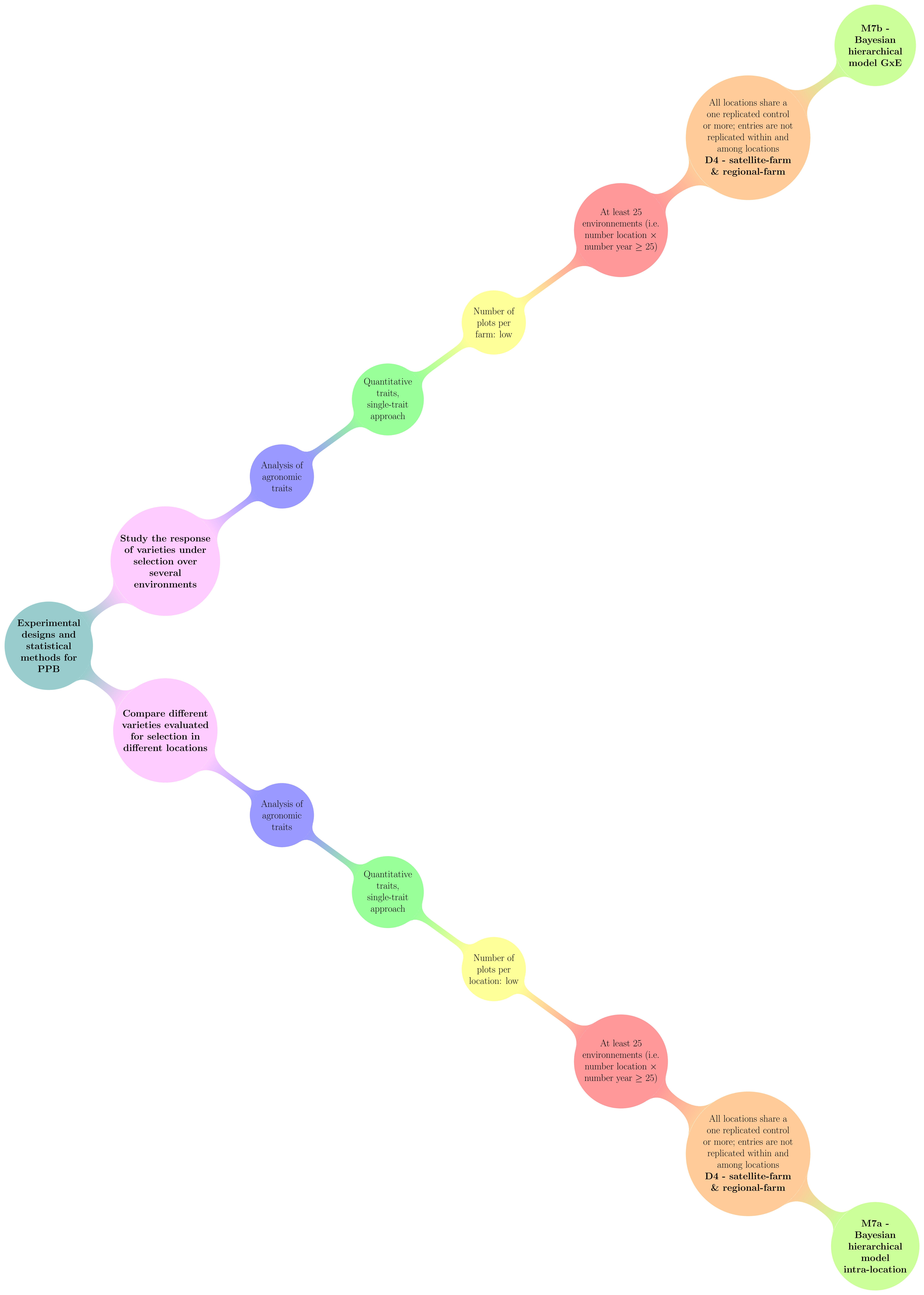
Figure 3.9: The decision tree where regional and satellite farms designs are used.
The R code to implement is the following :
As an example here, six designs are generated: four for satellite farms and two for regional farms.
p_case3_sf1 = design_experiment(
location = "Location-4",
year = "2016",
expe.type = "satellite-farm",
germplasm = paste("germ", c(1:6), sep = ":"),
controls = "germ:ctrl",
nb.controls.per.block = 2,
nb.blocks = 1,
nb.cols = 2)
p_case3_sf1 = p_case3_sf1$`satellite-farms`$design
p_case3_sf1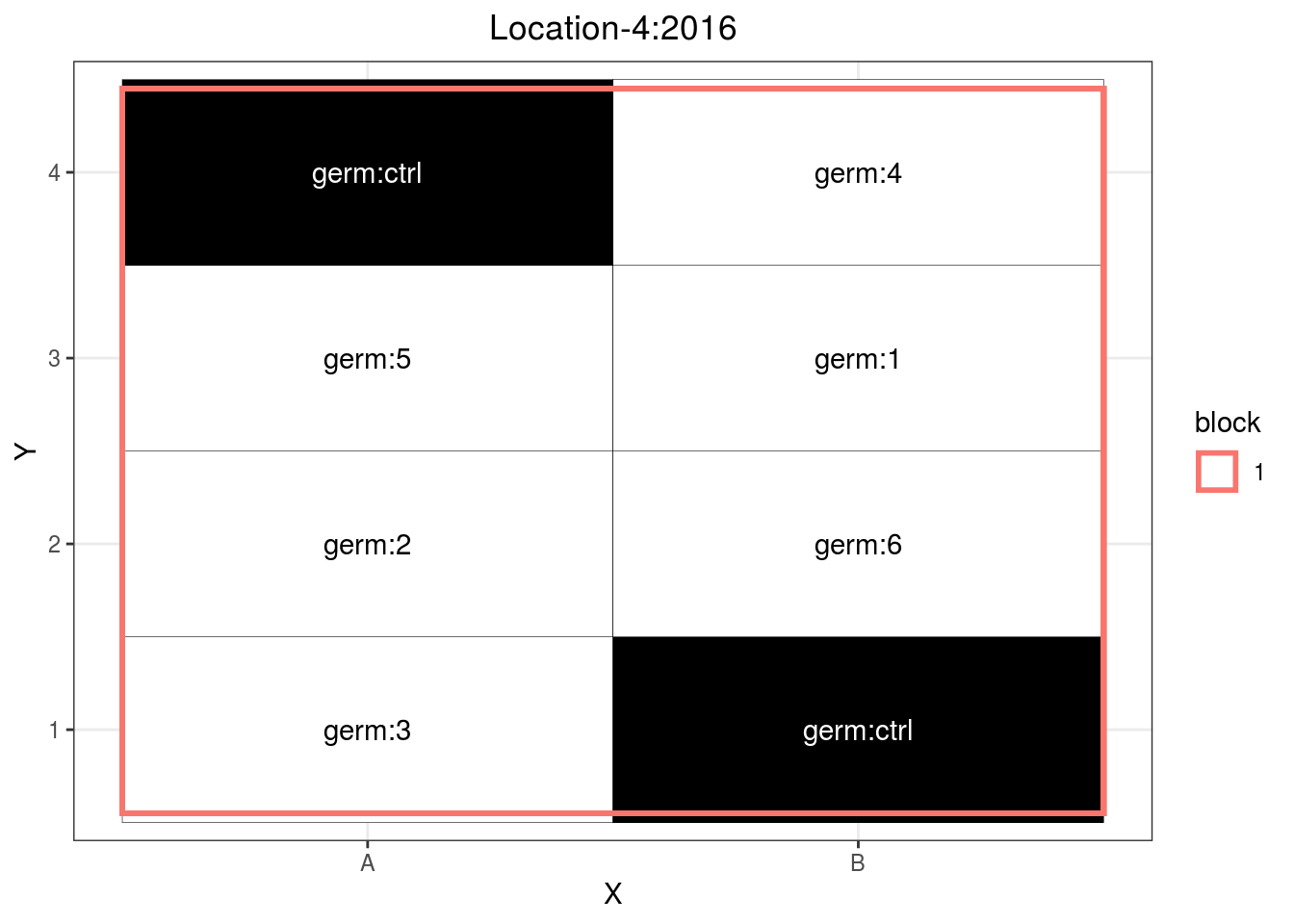
Figure 3.10: Example of a satellite farm design.
p_case3_sf2 = design_experiment(
location = "Location-5",
year = "2016",
expe.type = "satellite-farm",
germplasm = paste("germ", c(1:6), sep = ":"),
controls = "germ:ctrl",
nb.controls.per.block = 2,
nb.blocks = 1,
nb.cols = 2)
p_case3_sf2 = p_case3_sf2$`satellite-farms`$design
p_case3_sf2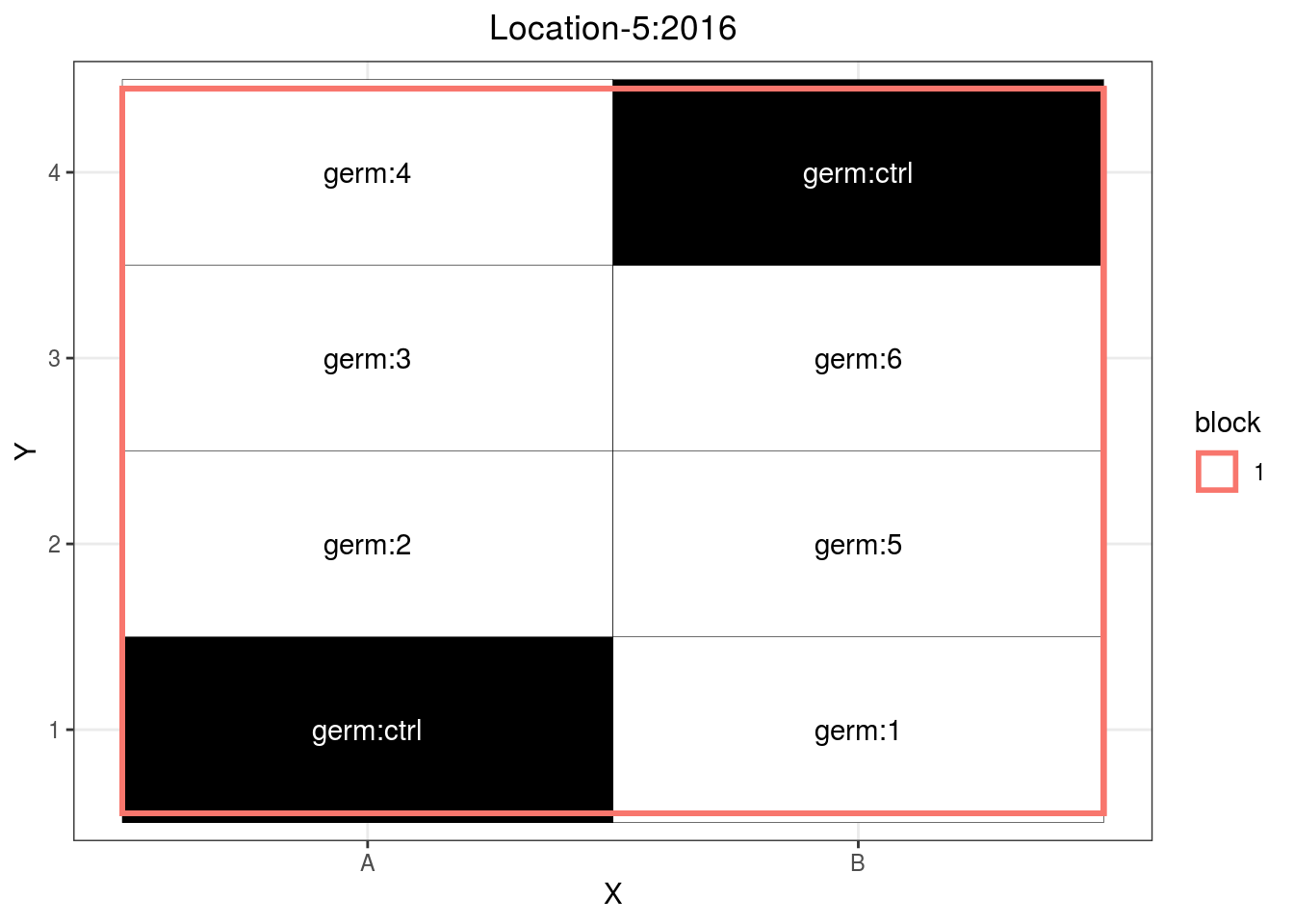
Figure 3.11: Example of a satellite farm design.
p_case3_sf3 = design_experiment(
location = "Location-6",
year = "2016",
expe.type = "satellite-farm",
germplasm = paste("germ", c(1:6), sep = ":"),
controls = "germ:ctrl",
nb.controls.per.block = 2,
nb.blocks = 1,
nb.cols = 2)
p_case3_sf3 = p_case3_sf3$`satellite-farms`$design
p_case3_sf3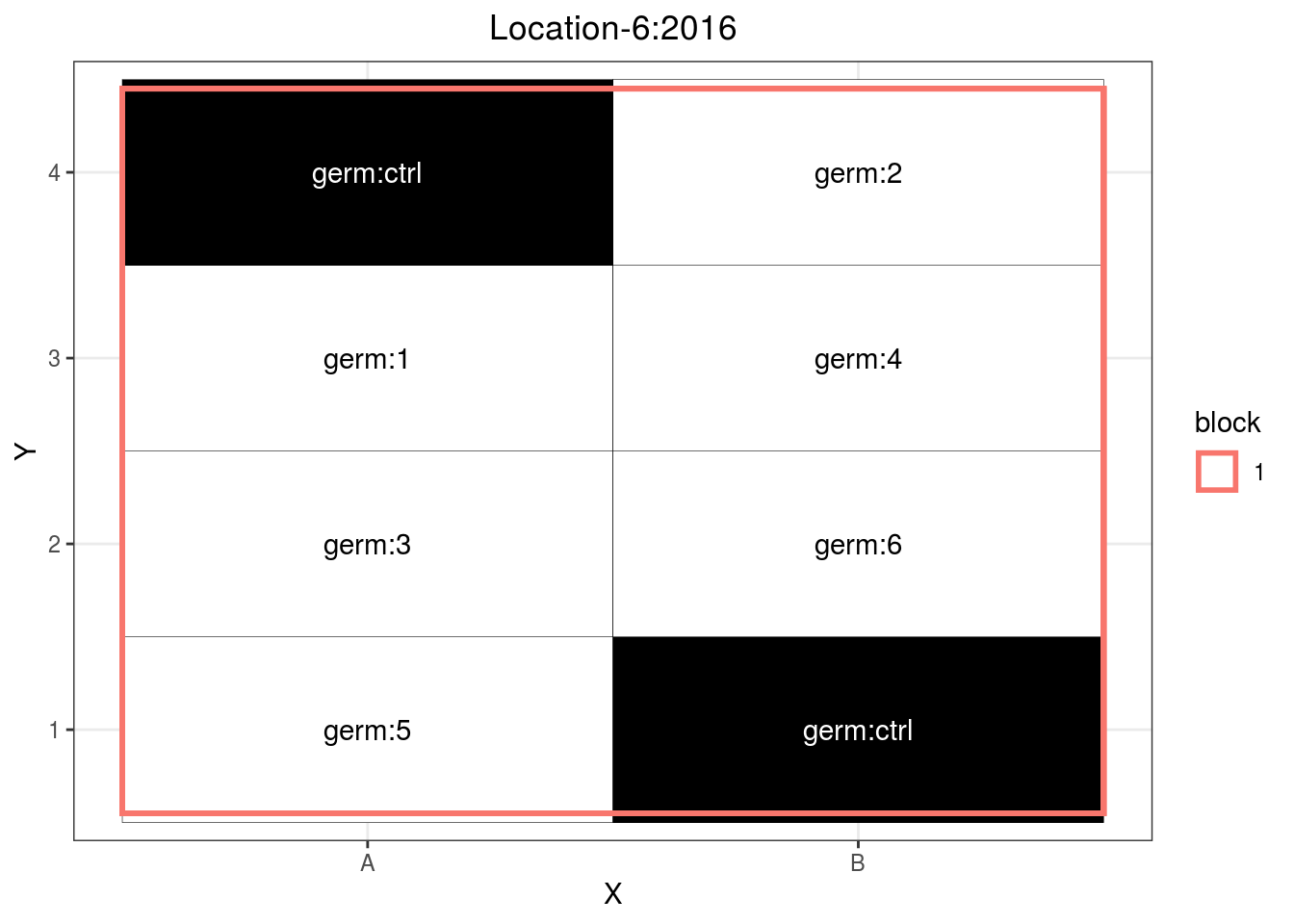
Figure 3.12: Example of a satellite farm design.
p_case3_rf1 = design_experiment(
location = "Location-7",
year = "2016",
expe.type = "regional-farm",
germplasm = paste("germ", c(1:16), sep = ":"),
controls = c("c1", "c2", "c3", "c4"),
nb.controls.per.block = 4,
nb.blocks = 2,
nb.cols = 4)
p_case3_rf1 = p_case3_rf1$`regional-farms`$design
p_case3_rf1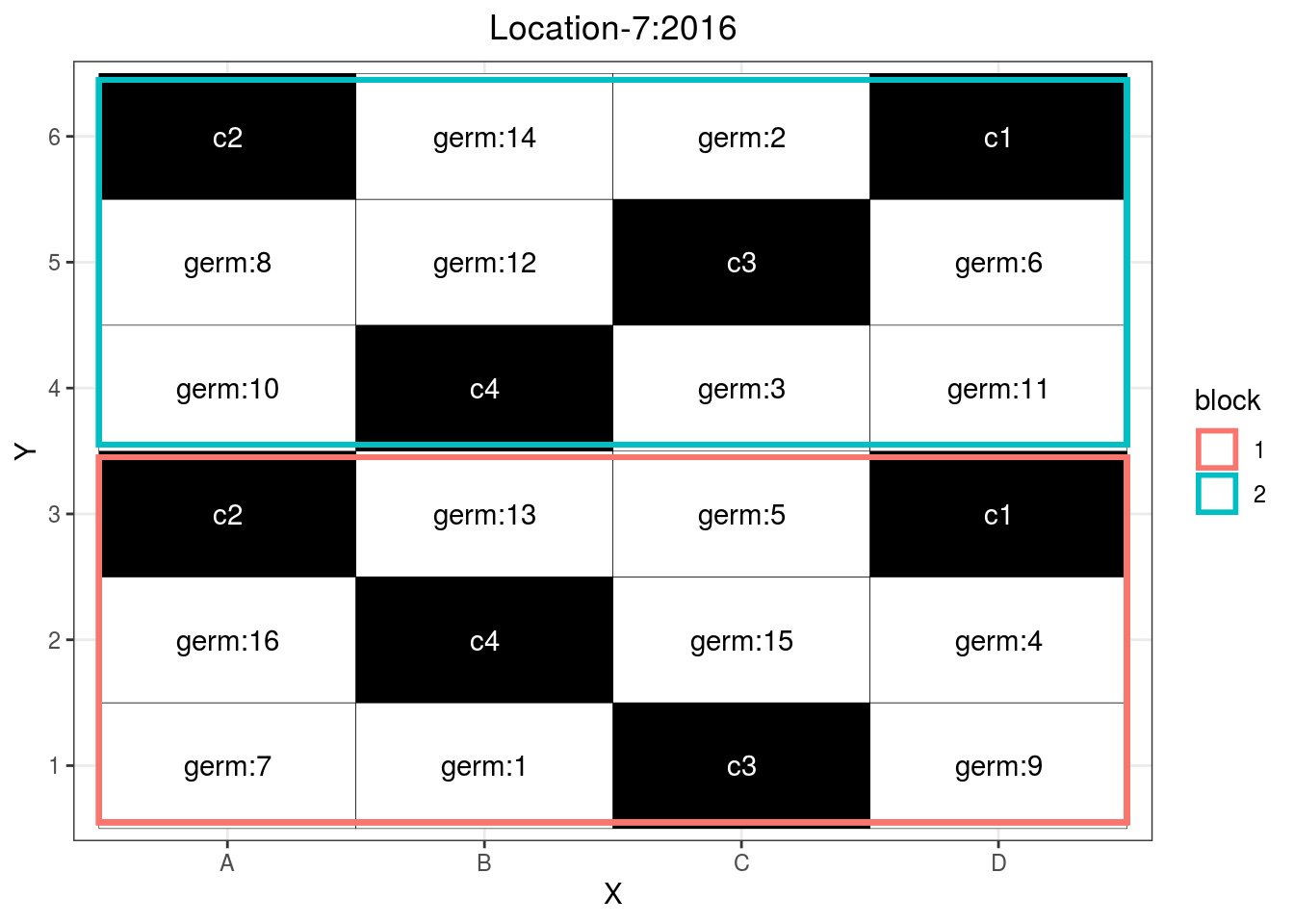
Figure 3.13: Example of a regional farm design.
p_case3_rf2 = design_experiment(
location = "Location-8",
year = "2016",
expe.type = "regional-farm",
germplasm = paste("germ", c(1:16), sep = ":"),
controls = c("c1", "c2", "c3"),
nb.controls.per.block = 3,
nb.blocks = 2,
nb.cols = 3)## Warning in place_controls(d, expe.type): Controls are missing in columns 3.
## You can rise nb.controls.per.block.## Warning in place_controls(d, expe.type): Controls are missing in rows 2.
## You can rise nb.controls.per.block.## Warning in place_controls(d, expe.type): Controls are missing in columns 2.
## You can rise nb.controls.per.block.## Warning in place_controls(d, expe.type): Controls are missing in rows 3.
## You can rise nb.controls.per.block.p_case3_rf2 = p_case3_rf2$`regional-farms`$design
p_case3_rf2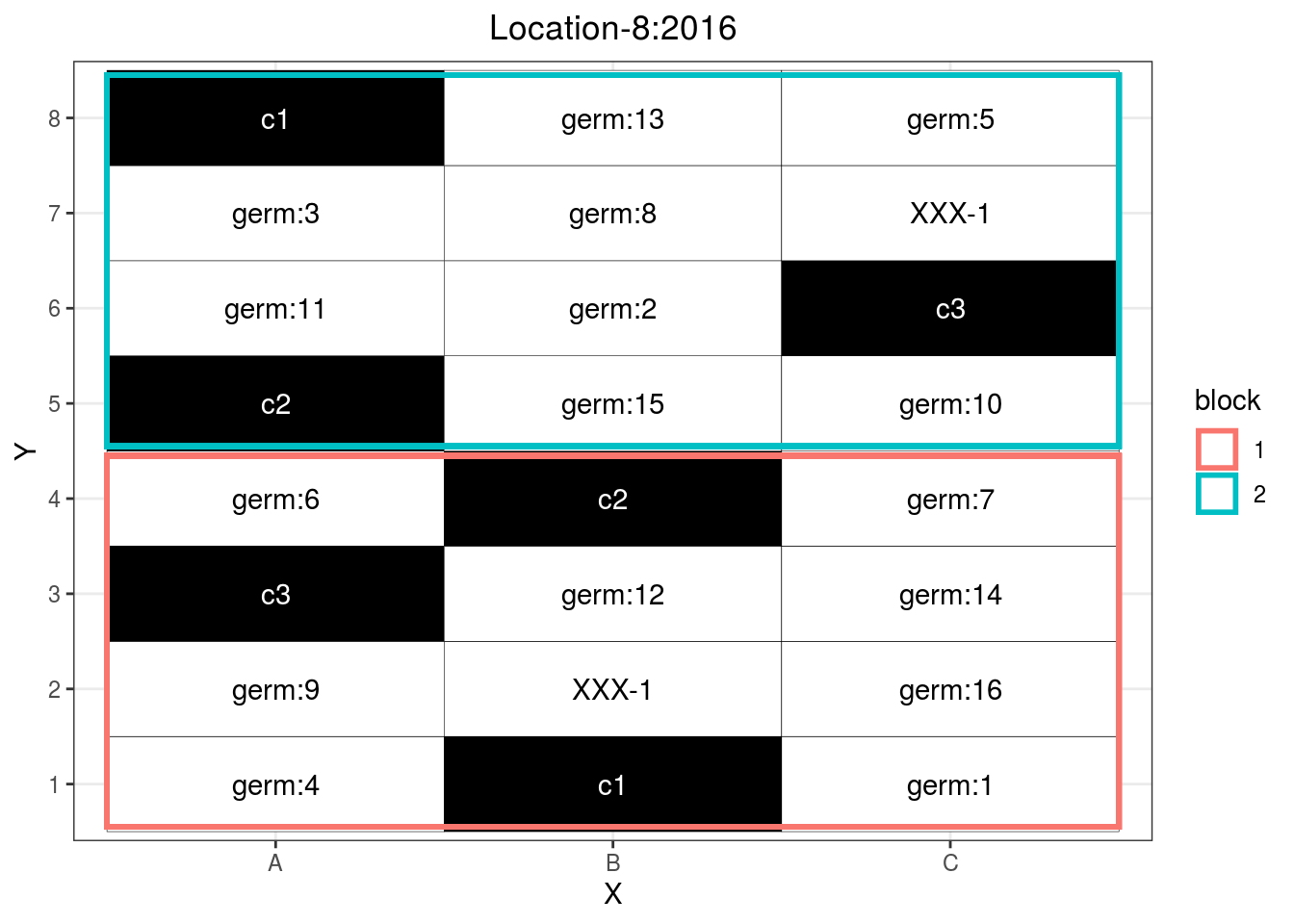
Figure 3.14: Example of a regional farm design.
If you have enought space and lots of seeds, you can adapt the satellite farm design with only one column. Each row beeing a sower width. Then you must be careful to have several sample for a given row to be as representative as possible.
p_case3_sf4 = design_experiment(
location = "Location-9",
year = "2016",
expe.type = "satellite-farm",
germplasm = paste("germ", c(1:6), sep = ":"),
controls = "C",
nb.controls.per.block = 2,
nb.blocks = 1,
nb.cols = 1)
p_case3_sf4 = p_case3_sf4$`satellite-farms`$design
p_case3_sf4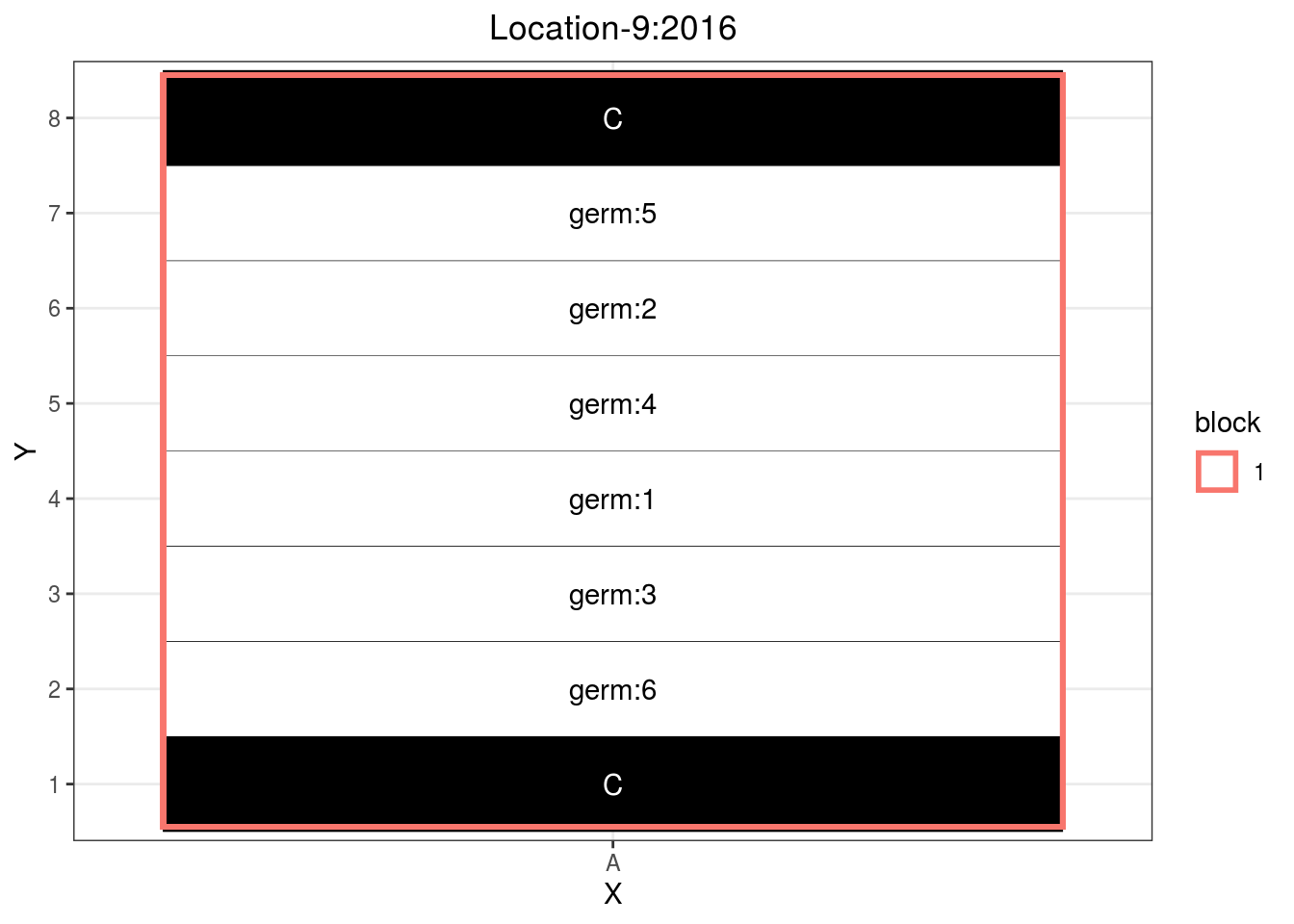
Figure 3.15: Example of a satellite farm design.
There some constraints regarding expe.type = "satellite-farm":
- if
nb.entries > 10, a warning message recommands to have less than 10 germplasms or to chooseexpe.type = "regional-farm". - There are two controls per block
- There is one block
- There are maximum two columns
For expe.type = "regional-farm", there is a warning message if controls are missing in rows or columns.
It is better to catch as much as possible of the trial variation.
If there are less than 2 blocks, an error message is returned.